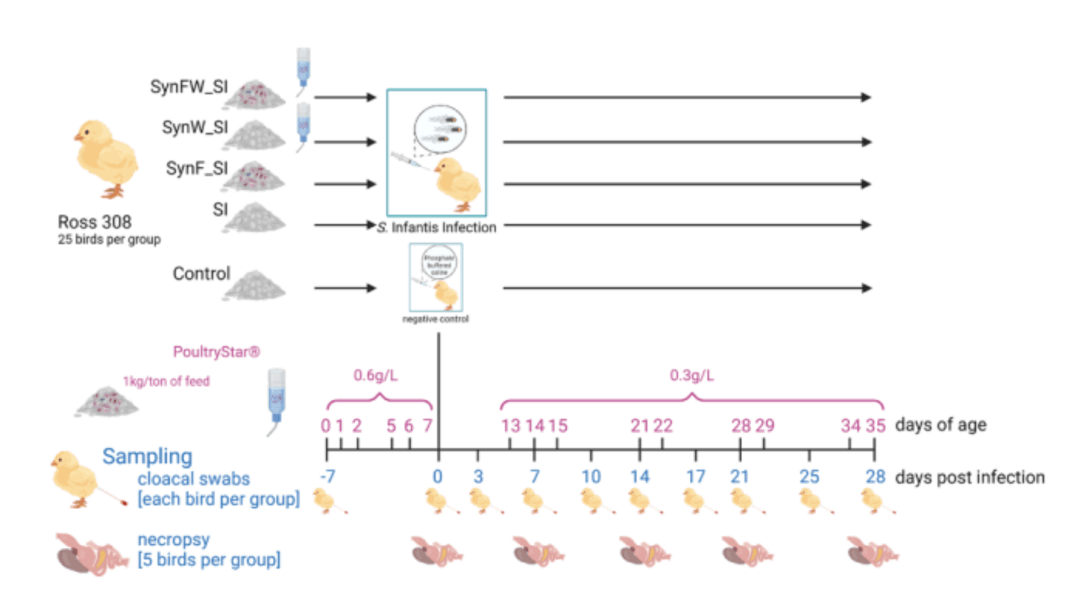
Differential effects of synbiotic delivery route (Feed, water, combined) in broilers challenged with Salmonella Infantis
Victoria Drauch et al. (2025)
Salmonella Infantis presents a persistent and multi-drug-resistant threat to poultry production. This study evaluated the
impact of a S. Infantis infection in broiler chickens across various parameters, including organ colonization, gut microbiota, and immune function. Our findings provide valuable guidance for
optimizing synbiotic use in commercial poultry management, enabling enhanced pathogen control and improved gut health.
You can read the full article on ScienceDirect! https://www.sciencedirect.com/science/article/pii/S0032579125001270?via%3Dihub