ABOUT 3D'OMICS

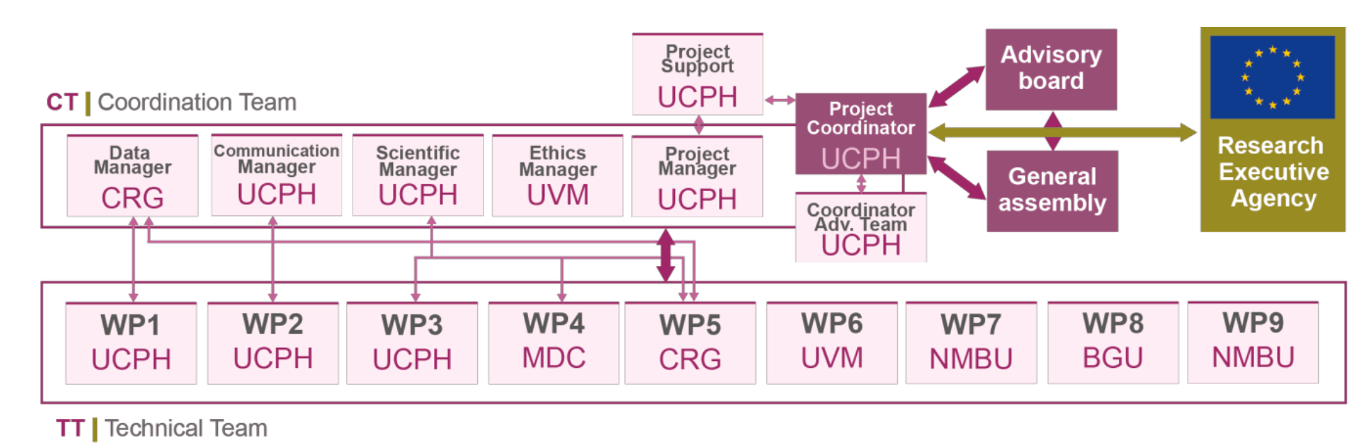
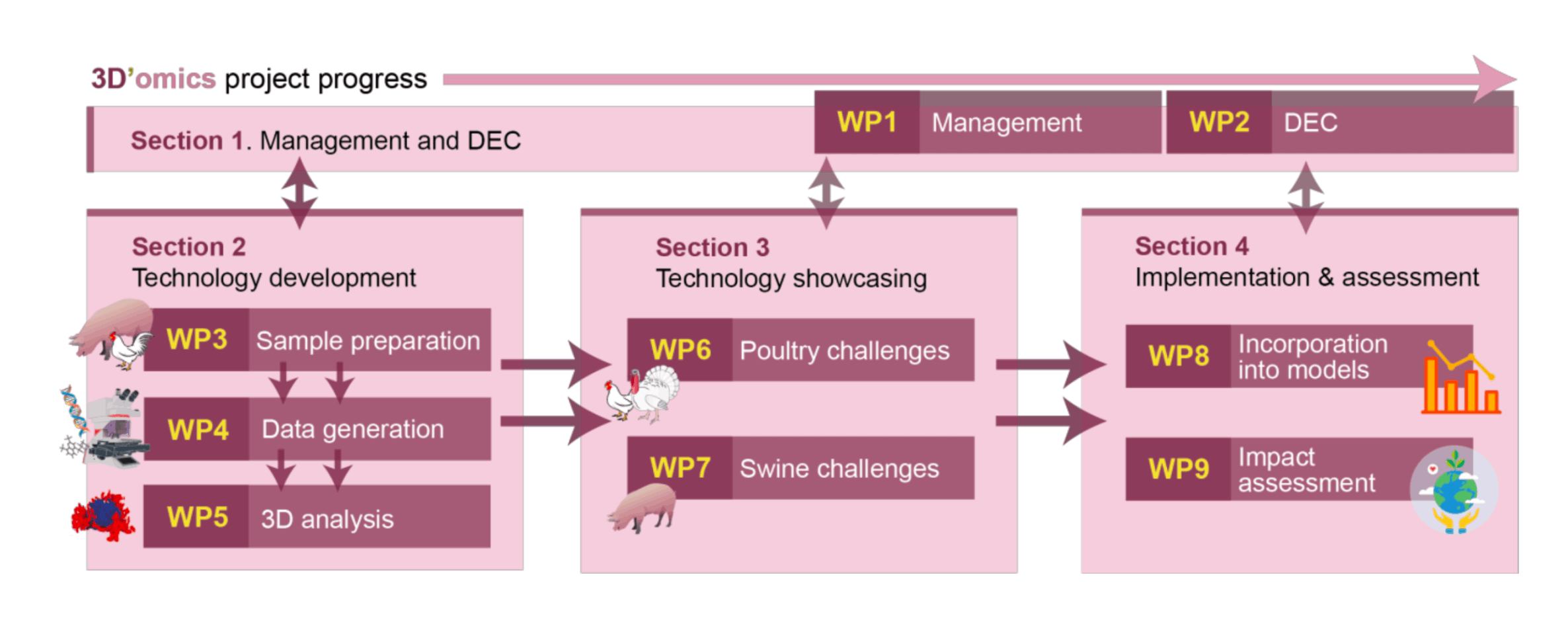
WP1: Project Management
Lead: UCPH
Management of the overall progress of the project through efficient scientific, administrative, financial, ethical, and data management. Guarantees that internal and EU reporting is performed adequately, makes certain that the project complies with ethical legislation, and ensures tasks and objectives are fulfilled efficiently within budget and on time.
WP2: Dissemination, Exploitation, and Communication (DEC)
Lead: UCPH
Communication of 3D’omics to relevant stakeholders and the general public, and promotion of new collaborative interactions with international projects to maximise the impact of 3D’omics technology.
WP3: Sample Preparation
Lead: UCPH
Development of optimal procedures for sampling, fixating, slicing, and micro-dissecting intestinal sections containing epithelial tissue and microbial communities. Generation of biological samples for the in vitro work in WP6/7, and creation of preliminary evidence for improving designs of trials in WP6/7.
WP4: Data Generation
Lead: MDC
Development of laboratory protocols and workflows to generate multi-omic data from intestinal microsections from different sample types.
WP5: Data Analysis and Visualisation
Lead: CRG
Development of the analytical procedures required to generate and visualise the 3D reconstruction models from 3D’omic data, and develop mechanistic and deep learning approaches to understand omic interactions between microbes and animals and predict phenotypic/performance outcomes.
WP6: Poultry Health Challenges
Lead: UVM
Showcasing the 3D’omics technology in addressing production challenges associated with diverse types of pathogens in poultry.
WP7: Swine Nutritional Challenges
Lead: NMBU
Showcasing the 3D’omics technology in addressing production challenges associated with nutrition in swine.
WP8: Incorporation into Models
Lead: BGU
Providing guidelines and expectations to implement 3D’omics data in genetic evaluation and phenotypic assessment procedures in industrial breeding and feeding programmes.
WP9: Technology Impact Assessment
Lead: NMBU
Assessment of the technical, social, and economic impact of the 3D’omics technology.
ABOUT 3D'OMICS